Molecular Screening Platform Kissf
Presentation
The specialized inhibitor screening platform, KISSf (Kinase Inhibitor Specialized Screening) offers a medium-throughput evaluation service for the bioactivity of chemical compounds. The analyzes carried out on a selected panel of protein kinases, allow the evaluation of the therapeutic properties of the molecules of interest. KISSf’s experience has made it possible to assess and highlight numerous compounds involved in cellular mechanisms and more particularly in mitosis and programmed cell death. The work of the platform, oriented in the field of biology and health, therefore aims to highlight future drug candidates.
The platform’s mission is therefore to produce new kinase targets and to develop enzymatic or interaction tests in order to be able to carry out molecular screening campaigns. Her benefits from expertise in the expression of recombinant protein in insect cells, and from an automated protein purification chain.
Field of study: Protein kinases (Characterization of Protein kinase Inhibitors)
Protein kinases are involved in the regulation of numerous cellular processes, often in response to an external stimulus. This family of enzymes has become one of the most important suppliers of drug targets. The number of approved small-molecule protein kinase inhibitors (PKIs) continues to grow and more then 50+ drugs have reached the US market, 85% of which are used for the treatment of malignancies. More than 200 orally effective PKIs are currently in clinical trials worldwide (a complete and updated listing of PKIs in clinical trials can be found at www.icoa.fr/pkidb/
Natural or synthetic chemical compounds and also semi-purified fractions of natural extracts can be analyzed on KISSf screening facility (Roscoff) to discover new PKIs. A structure-activity study can be performed to identify a lead compound. The binding mode of the hit compound to its kinase target can also be evaluated by an ATP competition assay.
Scientific expertise: Protein Kinase assays
Kinase assay:
Kinase enzymatic activities were assayed in 384-well plates using the ADP-GloTM assay kit (Promega, Madison, WI). This assay is a luminescent ADP detection assay that provides an homogeneous and high-throughput screening method to measure kinase activity by quantifying the amount of ADP produced during a kinase reaction.
Reactions were carried out in a final volume of 6 µl for 30 min at 30°C in appropriate kinase buffer, with either protein or peptide as substrate in the presence of 10µM ATP. After that, 6 µl of ADP-GloTM Kinase Reagent was added to stop the kinase reaction. After an incubation time of 50 min at room temperature (RT), 12 µl of Kinase Detection Reagent was added for one hour at RT. The transmitted signal was measured using the Envision (PerkinElmer, Waltham, MA) microplate luminometer and expressed in Relative Light Unit (RLU). Kinase activities are expressed in % of maximal activity, i.e. measured in the absence of inhibitor.
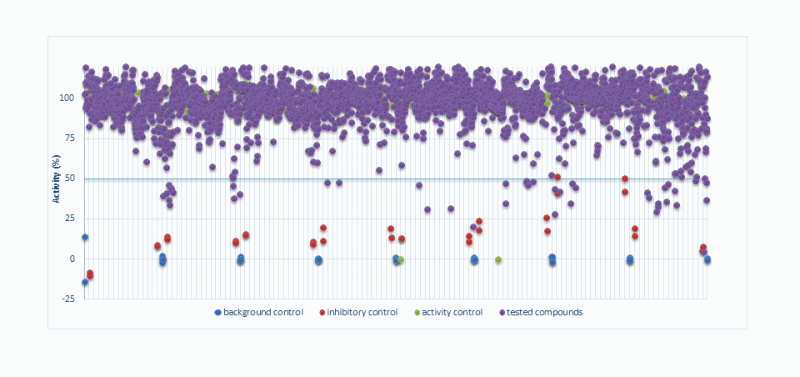
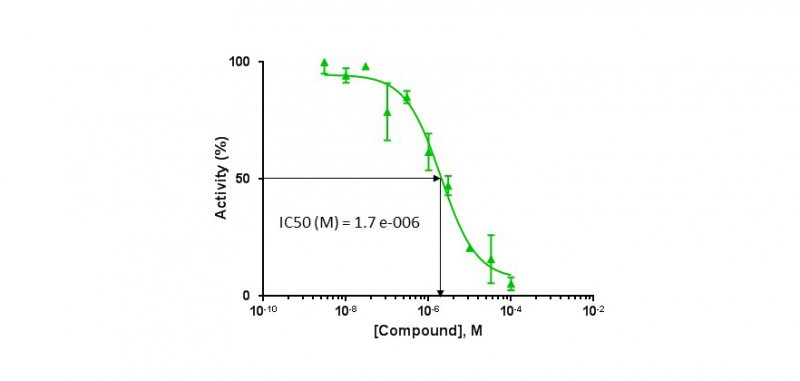
*(A) Primary kinase-based screening and * (B) determination of IC50
(A) Compounds are first tested at two concentrations (1 µM and 10 µM) against a panel of disease-related kinases. (B) Subsequently, compounds displaying more than 50% inhibition at 1 or 10 μM were next tested in duplicate over a wide range of concentrations (usually from 0.0003 to 10 μM) against the selected kinases and IC50 values were determined from the dose response curves using GraphPad Prism.
Primary Screening of chemical library
Determination of IC50
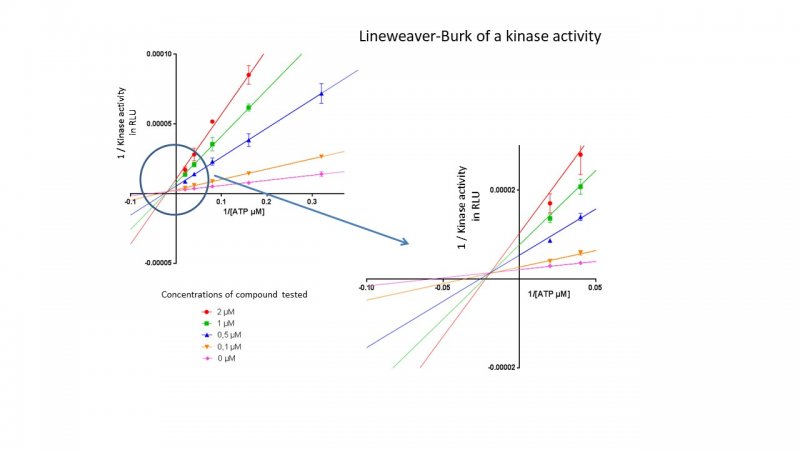
* ATP competition test by ADP-Glo method
To study the mechanism of inhibition of the hit compounds, KISSf performs kinetic experiments by varying both ATP levels and inhibitor concentrations to draw a double-reciprocal plot of the Michaelis-Menten equation (also known as the Lineweaver-Burk).
Kinase panel
Customers will select the kinases from the following list: 1. CDK2/CyclinA (CMGC), 2. CDK5/p25 (CMGC), 3. CDK9/CyclinT (CMGC), 4. CDK10/CyclinM (CMGC), 5. GSK3α (CMGC), 6. GSK3β (CMGC), 7. CLK1 (CMGC), 8. DYRK1A (CMGC), 9. PIM1 (CAMK), 10. CK1ε (CK1), 11. NEK6 (Other), 12. NEK7 (Other), 13. ABL1 (TK), 14. EGFR (TK), 15. VEGFR-2 (TK), 16. JAK3 (TK), 17. Haspin (Other), 18. RIPK3 (TKL), 19. PfGSK3 (from Plasmodium falciparum), 20. LmCK1 (from Leishmania major).
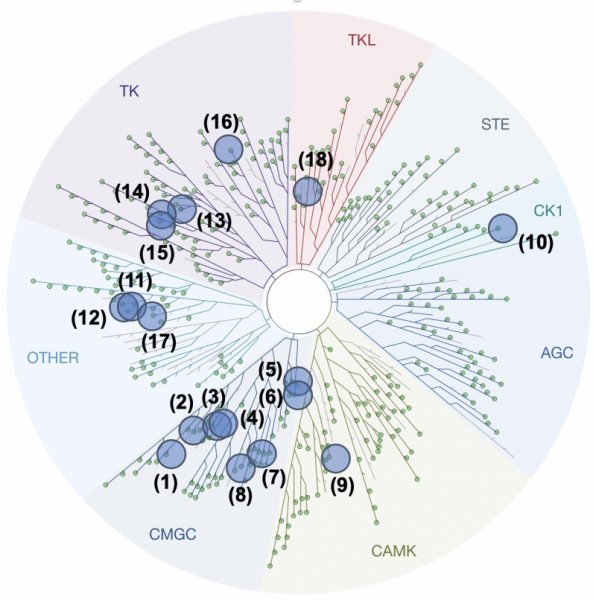
The panel of kinase available on KISSf reported on the representation of the human kinome.
The codes reported on this figure indicate the subclasses of protein kinases: CMGC for CDKs, MAP kinases, GSK, and CDK-like kinases; AGC for protein Kinase A, C, and G families (PKA, PKC, PKG); CAMK for Ca2+/calmodulin-dependent protein kinases; CK1, Cell/Casein Kinase 1; STE, STE Kinases (Homologs of yeast STErile kinases); TKL, Tyrosine Kinases-Like; TK, Tyrosine Kinases.
To validate each kinase assay, model inhibitors were used under the same conditions than the tested compounds. For example: Indirubin-3’-oxime for CDK2/CyclinA, CDK5/p25, CDK9/CyclinT, RnDYRK1A and MmCLK1; SGI-1776 for Pim1; Staurosporine from Streptomyces sp. for CK1ε; Tofacitinib for JAK3; Imatinib mesylate for ABL1; CHR-6494 for HASPIN; and GSK’872 for RIPK3.
How to request a quotation/ study:
If you want to collaborate with KISSf and add KISSf screening facility as partner of a grant request, please send us an email to: . If you want to submit chemical compounds for analysis against protein kinases, please fill-in the following google form and we will send you a quotation: https://docs.google.com/forms/d/1P8Q9HRjOfkACpVwOvdHCa7gmb26TVuxNmfgTWLi...
For all the requests from European academic labs that want to use our screening facility (robots, microplate readers, ...), our Institute is member of the EMBRC network and thus it is possible to welcome researchers, students and engineers: for details please use the link https://www.embrc-france.fr/fr
Articles published in 2019-2020
Cousseau A., Siano R., Probert I., Bach S. and Mehiri M. 2020. Marine Dinoflagellates as source of new bioactive structures. Studies in Natural Products Chemistry (Bioactive Natural Products), Vol 65 (1st Edition), ISBN 9780128179055. ELSEVIER SCIENCE PUBLISHERS – AMSTERDAM. In press.
Bouattour A., Fakhfakh M., Abid S., Paquin L., Le Guével R., Charlier T., Ruchaud S., Bach S., Bazureau J.-P. and Ammar H. 2020 Synthesis of new 2-phenylamino-4H-chromene-3-carbonitrile derivatives and their effects on tumor cell lines and against protein kinases. IJOC. In press.
Bach S., Colas P. et Blondel M. 2020. La levure modèle et outil… aussi pour la recherche thérapeutique. Médecine & Sciences. In press.
Ibrahima N., Bonnet P., Briona J.D., Peyrata J.F., Bignonc J., Levaiquec H., Josselin B., Robert T., Colas P., Bach S., Messaoudia S., Alamia M., Hamze A. 2020. Identification of a new series of Flavopiridol-like structures as kinase inhibitors with high cytotoxic potency. European Journal of Medicinal Chemistry. In press.
Benchekroun M., Ermolenko L., Tran M.Q.,Vagneux A., Nedev H., Delehouzé C., Souab M., Baratte B., Josselin B., Iorga B., Ruchaud S., Bach S.* and Al-Mourabit A.* 2020. Discovery of Simplified Benzazole Fragments Derived From The Marine Benzosceptrin B as Necroptosis Inhibitors Involving The Receptor Interacting Protein Kinase-1. European Journal of Medicinal Chemistry. In press.
Qhobosheane MA., Legoabe LJ., Josselin B., Bach S., Ruchaud S., Petzer J., Beteck R. 2020. Synthesis and evaluation of 7-azaindole derivatives bearing benzocycloalkanone motifs as protein kinase inhibitors [published online ahead of print, 2020 Mar 31]. Bioorg Med Chem. 2020;115468. doi:10.1016/j.bmc.2020.115468.
Robert T., Johnson J.L., Guichaoua R., Yaron T.M., Bach S., Cantley L.C. and Colas P. 2020. Development of a CDK10/CycM in vitro Kinase Screening Assay and Identification of First Small-Molecule Inhibitors. Front. Chem., 27 February 2020. DOI: 10.3389/fchem.2020.00147.
From simple quinoxalines to potent oxazolo[5,4-f]quinoxaline inhibitors of glycogen-synthase kinase 3 (GSK3). 2019. Lassagne F., Duguépéroux C., Roca C., Pérez C., Martinez A., Baratte B., Robert T., Ruchaud S., Bach S., Erb W., Roisnel T. and Mongin F. Org. Biomol. Chem., DOI: 10.1039/C9OB02002K.
Nascimento da Cruz A.C., Brondani D.J., I´talo de Santana T., Oliveira da Silva L., da Oliveira Borba E.F., de Faria A.R., Ferreira Cavalcanti de Albuquerque J., Piessard S., Matos Ximenes R., Baratte B., Bach S., Ruchaud S., Bezerra Mendonça Junior F.J., Bazin M.-A., Montenegro Rabello M., Hernandes M.Z., Marchand P. and Gonçalves da Silva T. 2019. Biological Evaluation of Arylsemicarbazone Derivatives as Potential Anticancer Agents. Pharmaceuticals, 12, 169.
Mokhtari Brikci-Nigassa N., Nauton L., Moreau P., Mongin O., Duval R., Picot L., Thiéry V., Souab M., Ruchaud S., Bach S., Guevel R., Bentabed-Ababsa G., Erb W., Roisnel T., Dorcet V., Mongin F. 2019. Functionalization of 9-thioxanthone at the 1-position: from arylamino derivatives to [1]benzo(thio)pyrano[4,3,2-de]benzothieno[2,3-b]quinolines of biological interest, Bioorganic Chemistry, 103347, doi.org/10.1016/j.bioorg.2019.103347.
Nguyen TN., Feizbakhsh O., Sfecci E., Baratte B., Delehouzé C., Garcia A., Moulin C., Colas P., Ruchaud S.*, Mehiri M.*, and Bach S. * 2019. Kinase-Based Screening of Marine Natural Extracts Leads to the Identification of a Cytotoxic High Molecular Weight Metabolite from the Mediterranean Sponge Crambe tailliezi. Mar Drugs. 2019 Oct 9;17(10). pii: E569. doi: 10.3390/md17100569.
Ziane S., Mazari M.M., Safer A.M., Sad El Hachemi Amar A., Ruchaud S., Baratte B. and Bach S. 2019. Comparison between Conventional and Nonconventional Methods for the Synthesis of Some 2-Oxazolidinone Derivatives and Preliminary Investigation of Their Inhibitory Activity Against Certain Protein Kinases. Russian Journal of Organic Chemistry. July 2019, Volume 55, Issue 7 (1061–1069). doi.org/10.1134/S1070428019070248.
Sartini S, Levati E., Maccesi M., Guerra M., Spadoni G., Bach S., Benincasa M., Scocchi M., Ottonello S., Rivara S. and Montanini B. 2019. New Antimicrobials Targeting Bacterial RNA Polymerase Holoenzyme Assembly Identified with an in Vivo BRET-Based Discovery Platform. ACS Chem Biol. 2019 Jul 30. doi: 10.1021/acschembio.9b00178.
Khaldoun K., Safer A., Boukabcha N., Dege N., Ruchaud S., Souab M., Bach S., Chouaih A. and Saidi-Besbes S. 2019. Synthesis and evaluation of new isatin-aminorhodanine hybrids as PIM1 and CLK1 kinase inhibitors. Journal of Molecular Structure. 1192 (82-90). ISSN 0022-2860. doi.org/10.1016/j.molstruc.2019.04.122.
Zeinyeh W., Esvan Y. J., Josselin B., Baratte B., Bach S., Nauton L., Théry V., Ruchaud S., Anizon F., Giraud F. and Moreau P. 2019. Kinase inhibitions in pyrido[4,3-h] and [3,4-g]quinazolines: Synthesis, SAR and molecular modeling studies. Bioorganic & Medicinal Chemistry. ISSN 0968-0896. doi.org/10.1016/j.bmc.2019.04.005.
Oukoloff K., Coquelle N., Bartolini M., Naldi M., Le Guevel R., Bach S., Josselin B., Ruchaud S., Catto M., Pisani L., Denora N., Iacobazzi R.M., Silman I., Sussman J.L., Buron F., Colletier J.-P., Jean L., Routier S. and Renard P.-Y. 2019. Design, biological evaluation and X-ray crystallography of nanomolar multifunctional ligands targeting simultaneously acetylcholinesterase and glycogen synthase kinase-3. European Journal of Medicinal Chemistry, doi.org/10.1016/j.ejmech.2018.12.063.
Motuhi S.-E., Feizbakhsh O., Foll-Josselin B., Baratte B., Delehouzé C., Cousseau A., Fant X., Bulinski J.C., Payri C.E., Ruchaud S., Mehiri M.* and Bach S.* 2019. Neurymenolide A, a Novel Mitotic Spindle Poison from the New Caledonian Rhodophyta Phacelocarpus neurymenioides. Mar. Drugs, 17, 93.
Tazarki H., Zeinyeha W., Esvana, Y.J., Snapp K., Chatterjee D., Schröder M., Joerger A. C., Khiarib J., Josselin B., Baratte B., Bach S., Ruchaud S., Anizona F., Giraud F. and Moreau P. 2019. New pyrido[3,4-g]quinazoline derivatives as CLK1 and DYRK1A inhibitors: synthesis, biological evaluation and binding mode analysis. European Journal of Medicinal Chemistry, https://doi.org/10.1016/j.ejmech.2019.01.052.
Masłyk, M., Janeczko, M., Martyna, A., Czernik, S., Tokarska-Rodak, M., Chwedczuk, M., Foll-Josselin, B., Ruchaud, S., Bach, S., Demchuk, O.M. and Kubiński, K. 2019. The Anti-Candida albicans Agent 4-AN Inhibits Multiple Protein Kinases. Molecules, 24, 153. doi.org/10.3390/molecules24010153.2019-2020:








