


ORIGIN, SPREAD AND EVOLUTION OF ANTIBIOTIC RESISTANCE GENES IN VIBRIOS
We discovered that resistance to multiple antibiotics is common in V. crassostreae strains isolated from the environment. Yet, our preliminary work shows that we ignore which are the ARG, how they are transferred and why they are kept in natural populations. Our first hypothesis is that environmental vibrios are reservoirs of ARGs, because these genes tend to be genetically linked with those involved in survival against toxic molecules present in the environment. For example, oysters naturally bio-accumulate heavy metals and selection for genes allowing bacteria to cope with them favor the acquisition of antibiotic resistance. Our second hypothesis is that oysters are hotspots of Horizontal Gene Transfer (HGT). This hypothesis is supported by the observation that conjugative transfer related genes are highly induced in V. crassostreae during oyster colonization (Rubio 2019).
To explore these hypotheses, we will study the mechanisms of HGT from a comparative genomics perspective, the ARG revealed by association genetics from a functional perspective, and the selection of these genes using both functional and population genetics approaches.
This project will be submitted for funding at the ANR PRC 2020 and will hire a PhD student for 3 years.
CollaboratorS
https://research.pasteur.fr/fr/team/microbial-evolutionary-genomics/
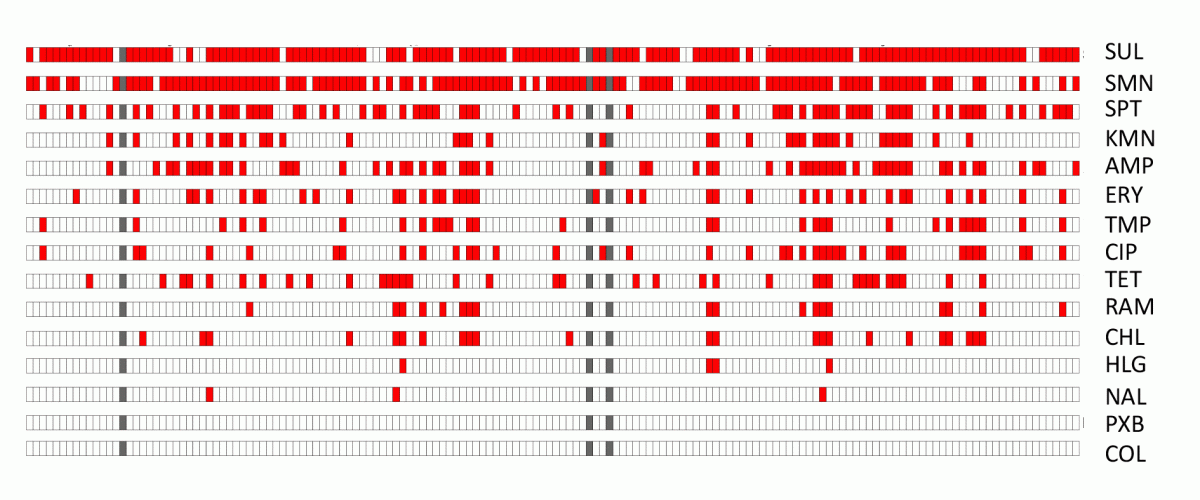
Antibiotic resistance of V. crassostreae environmental isolates. Diffusion in Muller Hinton agar, in red no inhibition, i.e. resistance. SUL, sulfamid; SMN, streptomycin; SPT, spectomycin; KMN, kanamycin; AMP, ampicillin; ERY, erythromycin; TMP, trimethoprim; CIP, ciprofloxacin; TET, tetracyclin, RAM, rifampicin; CHL, chloramphenicol; HLG, gentamycin; NAL, nalidixic acid; PXB, polymixin; COL, colistin.
Links
[1] https://research.pasteur.fr/fr/team/microbial-evolutionary-genomics/