Yacine Badis
Associate professor, Sorbonne University
Equipe génétique des algues

Molecular Phycopathology & Genome editing
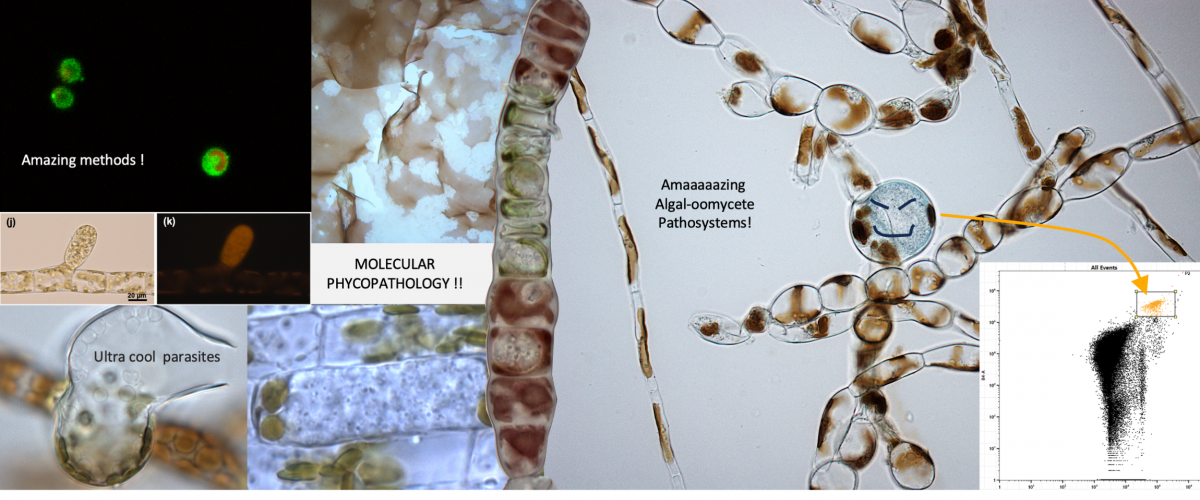
I develop macroalgal genome editing protocols to accelerate both fundamental and applied molecular research. I study the molecular actors of immune systems and algal-microbe interactions, using original biotrophic oomycete parasites as models. These oomycete parasites represent challenging and pervasive threats for seaweed hatcheries, my research thus aims to provide the seaweed sector with modern breeding molecular knoweledge and tools.
ONGOING PROJECTS
IMO-Drive: Intracellular Marine Oomycetes to DRIVE the discovery of brown algal immune systems (Principal investigator, funded by ANR JCJC 2024-2028 & Sorbonne Emergence)
PHABB: PatHogens of Algae for Biocontrol and Biosecurity (DC8): Functional and evolutionary genomics of brown algal immune-related genes for the future breeding of pathogen resistance in brown algae (HORIZON–MSCA-2022-DN) Apply until may 15th 2024 at https://mcam.mnhn.fr/en/phd-projects-within-phabb-application-guidelines-6691
BOOM-Drive: Biotrophic Oomycetes as DRIVErs for the elucidation of macroalgal immune systems (Principal Investigator, Sorbonne University kickstart funds 2022-2024)
HALOGENE: Deciphering the roles of HALOgen metabolism in a brown algal model through functional GENomics (WP1 lead, WP3 Co-lead)
Publications
Luthringer R, Raphalen M, Guerra C, Colin S, Martinho C, Hoshino M, Badis Y, Lipinska AP, Haas F, Barrera-Redondo J, Alva V, Coelho SM (2024). Repeated co-option of HMG-box genes for sex determination in brown algae and animals. Science 383, eadk5466 (2024). https://doi.org/10.1126/science.adk5466
Zuccarello G, Gachon CMM, Badis Y, Murúa P, Garvetto A, Kim GH (2024). Holocarpic oomycete parasites of red algae are not Olpidiopsis, but neither are they all Pontisma or Sirolpidium (Oomycota). Algae 2024; 39(1): 43-50.https://doi.org/10.4490/algae.2024.39.3.8
Badis Y, Scornet D, Harada M, Caillard C, Godfroy O, Raphalen M, Gachon CMM, Coelho SM, Motomura T, Nagasato C, Cock JM (2021). Targeted CRISPR-Cas9-based gene knockouts in the model brown alga Ectocarpus. New Phytologist Sep;231(5):2077-2091 https://doi.org/10.1111/nph.17525
Kloareg B, Badis Y, Cock JM, Michel G. Role and Evolution of the Extracellular Matrix in the Acquisition of Complex Multicellularity in Eukaryotes: A Macroalgal Perspective. Genes. 2021; 12(7):1059. https://doi.org/10.3390/genes12071059
Badis Y, Han JW, Klochkova TA, Gachon CMM & Kim GH (2020). The gene repertoire of Pythium porphyrae (Oomycota) suggests an adapted plant pathogen tackling red algae. Algae 35(2): 133-144. https://doi.org/10.4490/algae.2020.35.6.4
Badis Y, Brakel J, Klochkova TA, Ostrowski M, Tringe SG, Kim GH & Gachon MM (2019). Hidden diversity in the oomycete genus Olpidiopsis is a potential hazard to red algal cultivation and conservation worldwide. European Journal of Phycology, 55:2, 162-171, https://doi.org/10.1080/09670262.2019.1664769
Badis Y, Klochkova TA, Strittmatter M, Garvetto A, Murua P, Sanderson JC, Kim GH, Gachon CM (2018); Novel species of the oomycete Olpidiopsis potentially threaten European red algal cultivation, Journal of Applied Phycology. https://doi.org/10.1007/s10811-018-1641-9