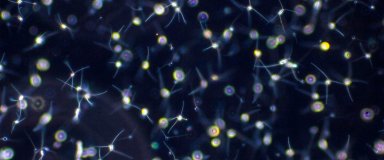
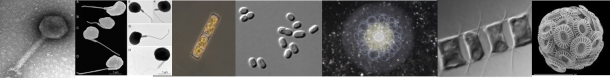
Structure and dynamics of planktonic communities
1) Address questions over different time scales using the plankton genomic observatory at Roscoff-Astan time-series combining high-throughput acquisition of advanced microscopic and multi-omics approaches (metabarcoding, metagenomics and metatranscriptomics) and fully capitalize on the strong expertise in taxonomy, imaging and functional diversity existing in the team and to a larger extent at the Station Biologique (e.g. ABIMS facility).
2) Explore geographical distribution of marine planktonic species (diversity and functions) using large scales datasets such as Tara-Oceans, Malaspina, Tara-Polar Circle, Tara-Pacific, Tara-Arctic II (2019-20) and Tara-Black Sea (2022) and other dedicated oceanographic cruises (ANR Tonga, Moose network, ANR Green Edge).
We combine culture-dependent approaches developed in close association with the Roscoff Culture Collection with meta-omics on the whole community and/or single-cell genomic data (i.e. single amplified genomes) to access the transcriptomes/genomes of uncultivated organisms and to obtain a more complete view of microbial community functions and dynamics. We also develop expertise databases such as the ones dedicated to protists [UniEuk, universal taxonomic framework for eukaryotes; PR2, 18S rRNA region; Aquasymbio, symbiotic associations], microalgae (PhytoREF, plastidial 16S rRNA), proteorhodopsin (PR)-containing photoheterotrophic bacteria (MicRhoDE, PR gene) and cyanobacteria (CyanoDB, petBgene).
The ECOMAP ambitions are:
1) to address questions over different time scales using the plankton genomic observatory at Roscoff-Astan time-series combining high-throughput acquisition of advanced microscopic and multi-omics approaches (metabarcoding, metagenomics and metatranscriptomics) and fully capitalize on the strong expertise in taxonomy, imaging and functional diversity existing in the team and to a larger extent at the Station Biologique (e.g. Mer & Plongée and platform facilities).
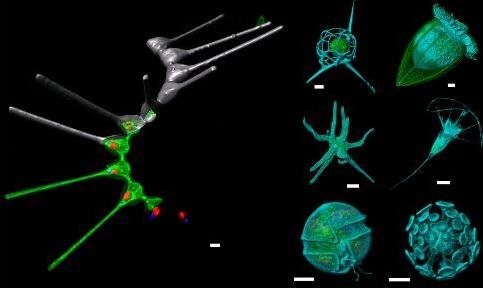
2) to explore geographical distribution of marine planktonic species (diversity and functions) using large scales datasets such as Tara-Oceans, Malaspina, Tara-Polar Circle, Tara-Pacific, Tara-Microbiomes, Plankton Planet, Bougainville) and other dedicated oceanographic cruises (ANR Tonga, Moose network, ANR Green Edge, ANR Apero).
We combine culture-dependent approaches developed in close association with the Roscoff Culture Collection with meta-omics on the whole community and/or single-cell genomic data (i.e. single amplified genomes) to access the transcriptomes/genomes of uncultivated organisms and to obtain a more complete view of microbial community functions and dynamics. We also develop expertise databases such as the ones dedicated to protists [UniEuk, universal taxonomic framework for eukaryotes; PR2, 18S rRNA region; Aquasymbio, symbiotic associations], microalgae (PhytoREF, plastidial 16S rRNA), proteorhodopsin (PR)-containing photoheterotrophic bacteria (MicRhoDE, PR gene) and cyanobacteria (CyanoMarks, petBgene).
Ongoing projects
- ANR PHENOMAP (2020-24)
- BOUGAINVILLE (2022-25)
- APERO (2022-26)
- BiOCEAN 5D (2022-26)
- FUTURE-OBS (2022-28)
- PLANKTON PLANET
- EMO-BON
- PIA AO EMBRC
We will be happy to share our expertise in the frame of large scale projects
Links
[1] http://www.sb-roscoff.fr/fr/observation/presentation/perimetrecarte-des-points-de-suivi/astan
[2] http://resomar.cnrs.fr/Base-de-donnee-Pelagos
[3] http://www.sb-roscoff.fr/fr/station-biologique-de-roscoff/services/moyens-a-la-mer-plongee-scientifique/
[4] http://www.sb-roscoff.fr/fr/station-biologique-de-roscoff/services/plateformes-technologiques/
[5] https://oceans.taraexpeditions.org/en/m/about-tara/les-expeditions/tara-oceans/
[6] http://www.expedicionmalaspina.es/
[7] https://oceans.taraexpeditions.org/jdb/lexpedition-tara-oceans-polar-circle/
[8] https://oceans.taraexpeditions.org/en/m/about-tara/les-expeditions/tara-pacific/
[9] https://fondationtaraocean.org/expedition/mission-microbiomes/
[10] https://planktonplanet.org/
[11] https://mission-bougainville.fr/
[12] http://www.agence-nationale-recherche.fr/en/anr-funded-project/?tx_lwmsuivibilan_pi2%5BCODE%5D=ANR-18-CE01-0016
[13] http://www.moose-network.fr/
[14] http://www.greenedgeproject.info/
[15] https://anr.fr/Project-ANR-21-CE01-0027
[16] http://roscoff-culture-collection.org/
[17] https://unieuk.org/
[18] https://github.com/pr2database/pr2database
[19] http://www.aquasymbio.fr/fr
[20] http://phytoref.sb-roscoff.fr/
[21] http://application.sb-roscoff.fr/micrhode/
[22] https://www.biocean5d.org/
[23] https://www.sorbonne-universite.fr/en/news/ocean-national-research-agency-supports-sorbonne-university-project
[24] https://www.embrc.eu/emo-bon
[25] https://www.embrc-france.fr/fr